Tools
As a lab, we release and periodically update all the tools we develop and use for our research. Our tools are open sourced, and we encourage you to reach out to us with any collaboration or issues. The goal is to produce high quality, clean tools that can be easily utilized by any research with minimal investment to learning how the tool works. Sometimes we offer a tool as a webserver for easier use, though standalone local version exist as well.
Our tools, curated databases, and webservers can be found below. If you use one of these please cite it! Our students and researchers work hard to make these tools available for easy use, give them a pat on the back.
Predictive models
 BANANABAsic NeurAl Network for bindding Affinity
BANANABAsic NeurAl Network for bindding AffinityMPNN for predicting binding affinity of small molecules to protein pockets.
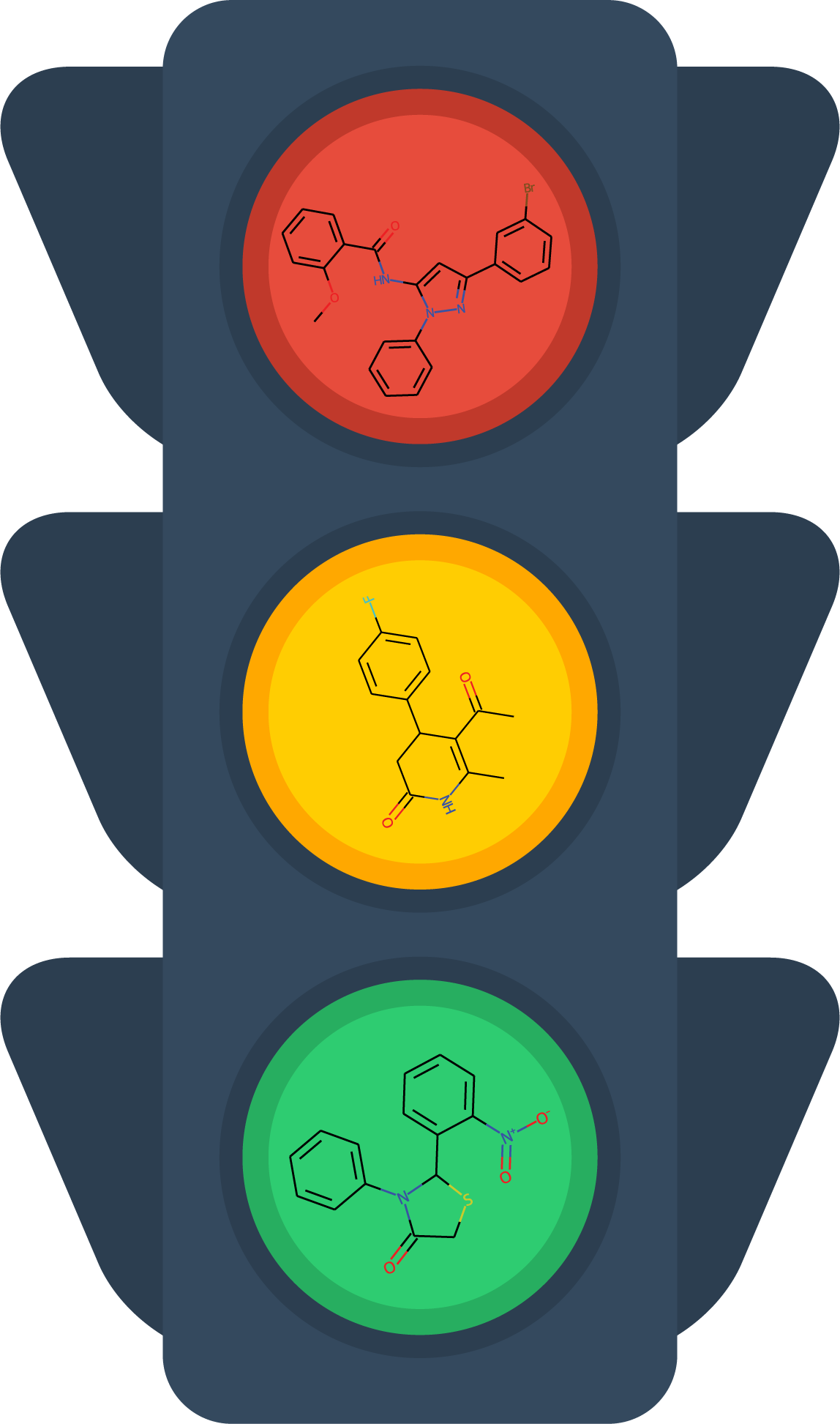 STOPLIGHTDrug-likeness Scoring Calculator
STOPLIGHTDrug-likeness Scoring CalculatorA webserver for generating a “goodness” score for early stage hits in drug discovery campaigns
 STopToxSystemic and Topical chemical Toxicity
STopToxSystemic and Topical chemical ToxicityA machine learning app to predict the battery of acute toxicity test classically known as “6-pack”
 PhaKinProPharmacokinetic Property calculator
PhaKinProPharmacokinetic Property calculatorA machine learning app to predict several human pharmacokinetic endpoint for small molecules
 LiabilityPredictor
LiabilityPredictorA machine learning app to predict if a small molecule is at risk of interfering/being a liability to an assay
 SALSAStructurally Aware Latent Space Auto-encoder
SALSAStructurally Aware Latent Space Auto-encoderA contrastive autoencoder for generating a latent space where chemicals of similar graph edit distance are near each other
Methods
Databases
 BigBind
BigBindA dataset built from CrossDocked and ChEMBL data
 SMACCSmall Molecule Antiviral Compound Collection
SMACCSmall Molecule Antiviral Compound CollectionA dataset a small molecules that have antiviral activity collected from ChEMBL.
Tools
 ExEmPLARExtracting, Exploring, and Embedding Pathways Leading to Actionable Research
ExEmPLARExtracting, Exploring, and Embedding Pathways Leading to Actionable ResearchA user-interface for exploring and analyzing biomedical knowledge graphs.
